Note
Go to the end to download the full example code
Blockwise coregistration#
Often, biases are spatially variable, and a “global” shift may not be enough to coregister a DEM properly.
In the Nuth and Kääb coregistration example, we saw that the method improved the alignment significantly, but there were still possibly nonlinear artefacts in the result.
Clearly, nonlinear coregistration approaches are needed.
One solution is xdem.coreg.BlockwiseCoreg, a helper to run any Coreg class over an arbitrarily small grid, and then “puppet warp” the DEM to fit the reference best.
The BlockwiseCoreg class runs in five steps:
Generate a subdivision grid to divide the DEM in N blocks.
Run the requested coregistration approach in each block.
Extract each result as a source and destination X/Y/Z point.
Interpolate the X/Y/Z point-shifts into three shift-rasters.
Warp the DEM to apply the X/Y/Z shifts.
import geoutils as gu
import matplotlib.pyplot as plt
import numpy as np
import xdem
Example files
reference_dem = xdem.DEM(xdem.examples.get_path("longyearbyen_ref_dem"))
dem_to_be_aligned = xdem.DEM(xdem.examples.get_path("longyearbyen_tba_dem"))
glacier_outlines = gu.Vector(xdem.examples.get_path("longyearbyen_glacier_outlines"))
# Create a stable ground mask (not glacierized) to mark "inlier data"
inlier_mask = ~glacier_outlines.create_mask(reference_dem)
plt_extent = [
reference_dem.bounds.left,
reference_dem.bounds.right,
reference_dem.bounds.bottom,
reference_dem.bounds.top,
]
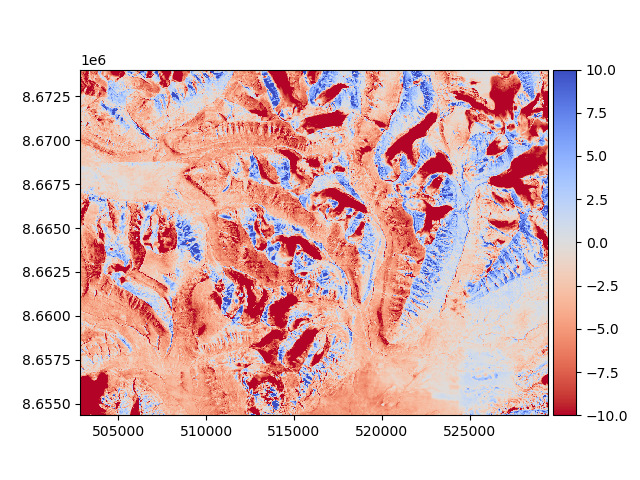
The DEM to be aligned (a 1990 photogrammetry-derived DEM) has some vertical and horizontal biases that we want to avoid, as well as possible nonlinear distortions. The product is a mosaic of multiple DEMs, so “seams” may exist in the data. These can be visualized by plotting a change map:
diff_before = reference_dem - dem_to_be_aligned
diff_before.plot(cmap="coolwarm_r", vmin=-10, vmax=10)
plt.show()
Horizontal and vertical shifts can be estimated using xdem.coreg.NuthKaab.
Let’s prepare a coregistration class that calculates 64 offsets, evenly spread over the DEM.
blockwise = xdem.coreg.BlockwiseCoreg(xdem.coreg.NuthKaab(), subdivision=64)
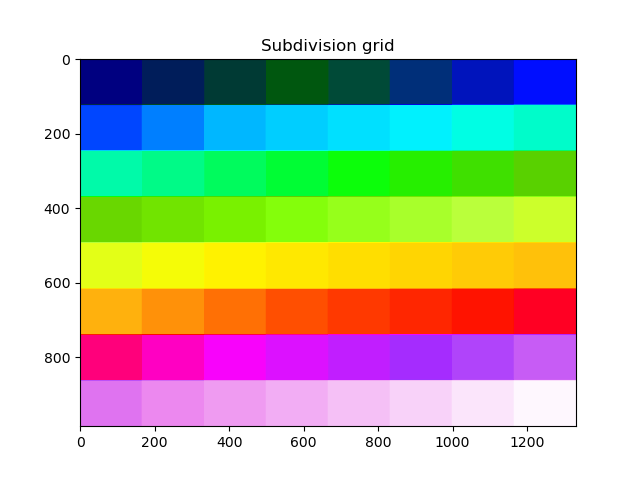
The grid that will be used can be visualized with a helper function. Coregistration will be performed in each block separately.
plt.title("Subdivision grid")
plt.imshow(blockwise.subdivide_array(dem_to_be_aligned.shape), cmap="gist_ncar")
plt.show()
Coregistration is performed with the .fit() method.
This runs in multiple threads by default, so more CPU cores are preferable here.
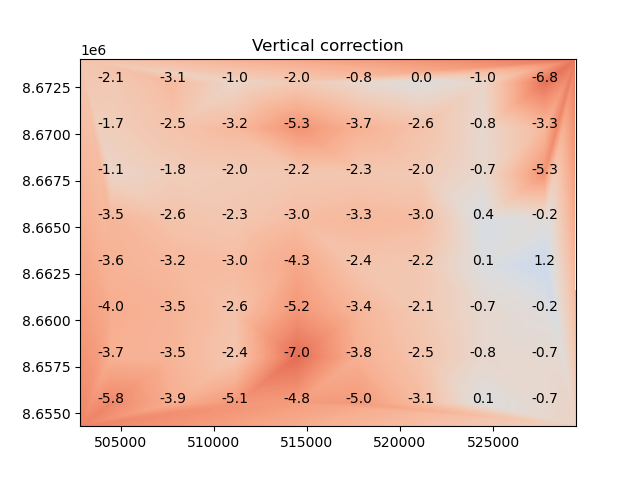
The estimated shifts can be visualized by applying the coregistration to a completely flat surface.
This shows the estimated shifts that would be applied in elevation; additional horizontal shifts will also be applied if the method supports it.
The xdem.coreg.BlockwiseCoreg.stats() method can be used to annotate each block with its associated Z shift.
z_correction = blockwise.apply(
np.zeros_like(dem_to_be_aligned.data), transform=dem_to_be_aligned.transform, crs=dem_to_be_aligned.crs
)[0]
plt.title("Vertical correction")
plt.imshow(z_correction, cmap="coolwarm_r", vmin=-10, vmax=10, extent=plt_extent)
for _, row in blockwise.stats().iterrows():
plt.annotate(round(row["z_off"], 1), (row["center_x"], row["center_y"]), ha="center")
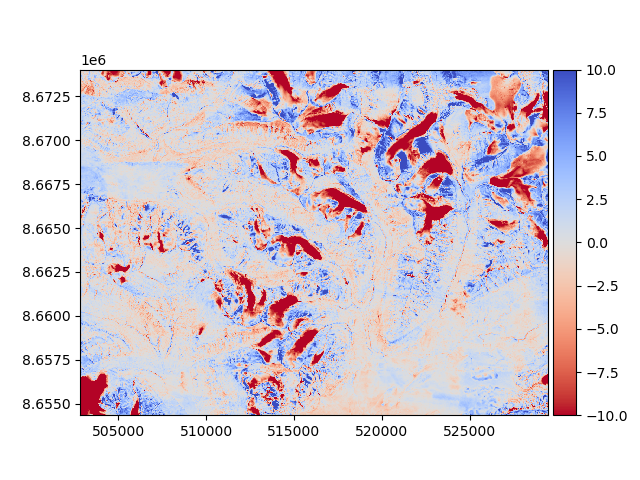
Then, the new difference can be plotted to validate that it improved.
diff_after = reference_dem - aligned_dem
diff_after.plot(cmap="coolwarm_r", vmin=-10, vmax=10)
plt.show()
We can compare the NMAD to validate numerically that there was an improvment:
print(f"Error before: {xdem.spatialstats.nmad(diff_before):.2f} m")
print(f"Error after: {xdem.spatialstats.nmad(diff_after):.2f} m")
Error before: 3.81 m
Error after: 2.24 m
Total running time of the script: (0 minutes 9.908 seconds)