Note
Go to the end to download the full example code
Bias correction with deramping#
(On latest only) Update will follow soon with more consistent bias correction examples.
In xdem, this approach is implemented through the xdem.biascorr.Deramp class.
For more information about the approach, see Deramping.
import geoutils as gu
import numpy as np
import xdem
Example files
reference_dem = xdem.DEM(xdem.examples.get_path("longyearbyen_ref_dem"))
dem_to_be_aligned = xdem.DEM(xdem.examples.get_path("longyearbyen_tba_dem"))
glacier_outlines = gu.Vector(xdem.examples.get_path("longyearbyen_glacier_outlines"))
# Create a stable ground mask (not glacierized) to mark "inlier data"
inlier_mask = ~glacier_outlines.create_mask(reference_dem)
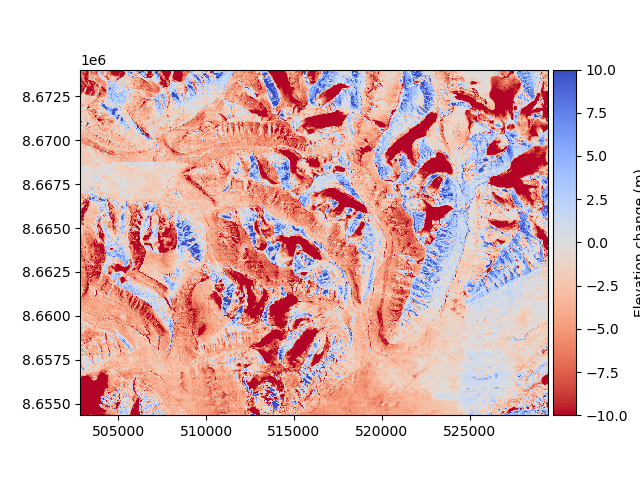
The DEM to be aligned (a 1990 photogrammetry-derived DEM) has some vertical and horizontal biases that we want to avoid. These can be visualized by plotting a change map:
diff_before = reference_dem - dem_to_be_aligned
diff_before.plot(cmap="coolwarm_r", vmin=-10, vmax=10, cbar_title="Elevation change (m)")
A 2-D 3rd order polynomial is estimated, and applied to the data:
Then, the new difference can be plotted.
diff_after = reference_dem - corrected_dem
diff_after.plot(cmap="coolwarm_r", vmin=-10, vmax=10, cbar_title="Elevation change (m)")
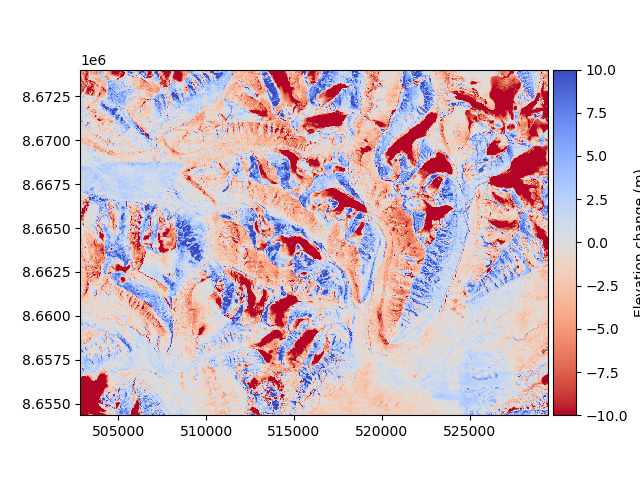
We compare the median and NMAD to validate numerically that there was an improvement (see Measures of central tendency and dispersion):
inliers_before = diff_before[inlier_mask]
med_before, nmad_before = np.median(inliers_before), xdem.spatialstats.nmad(inliers_before)
inliers_after = diff_after[inlier_mask]
med_after, nmad_after = np.median(inliers_after), xdem.spatialstats.nmad(inliers_after)
print(f"Error before: median = {med_before:.2f} - NMAD = {nmad_before:.2f} m")
print(f"Error after: median = {med_after:.2f} - NMAD = {nmad_after:.2f} m")
/home/docs/checkouts/readthedocs.org/user_builds/xdem/conda/latest/lib/python3.11/site-packages/numpy/core/fromnumeric.py:771: UserWarning: Warning: 'partition' will ignore the 'mask' of the MaskedArray.
a.partition(kth, axis=axis, kind=kind, order=order)
Error before: median = -2.33 - NMAD = 3.42 m
Error after: median = -0.20 - NMAD = 2.91 m
Total running time of the script: (0 minutes 2.080 seconds)